Molecular characterization of thermophilic Streptomyces using amplified 16s ribosomal DNA restriction analysis (ARDRA)
Keywords:
Streptomyces, PIB Win, Hae III, Eco R I, ARDRAAbstract
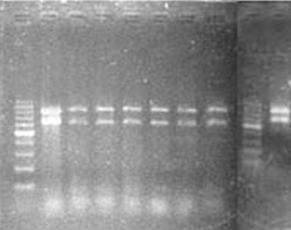
The present study aims to characterize the 19 thermophilic Streptomyces isolates on the basis of their restriction profile obtained by Amplified Ribosomal DNA Restriction Analysis (ARDRA) method. Nineteen thermophilic Streptomyces isolates, characterized and identified on the basis of cultural, morphological, physiological and biochemical characteristics using PIB Win (Probabilistic identification of Bacteria) software, have been further characterized by Amplified Ribosomal DNA Restriction Analysis method. DNA of all the 19 selected isolates and two reference Streptomyces cultures of MTCC (S. phaeochromogenes (MTCC 2614) and S. thermovulgaris (MTCC 1822) were isolated and the 16S rRNA gene was amplified using primer F1 (5'-AGAGTTTGATCITGGCTCAG-3’; I= inosine) and primer R5 (5'-ACGGITACCTTGTTACGACTT-3'). Two restriction endonuclease enzymes i.e. Eco R I and Hae III were used for restriction digestion. The 16S rDNA were amplified by PCR using specific primers and the amplified product were cleaved separately by both endonucleases, yielding 4 to 5 bands by Hae III and 2 to 3 bands by Eco R I. Digestion with two endonucleases indicate that Hae III enzyme is better than Eco R I for differentiating Streptomyces isolates.
References
Bauer, A.W., Kirby, W.M., Sherris, J.C., Turk, M., 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Path., 45, 493–496.
Berdy, J., 1995. Are actinomycetes exhausted as a source of secondary metabolites? In Proceedings of the 9th International Symposium on the Biology of Actinomycetes, part I. New York: Allerton. 3–23.
Beyazova, M., Lechevalier, M.P., 1993. Taxonomic utility of restriction endonuclease fingerprinting of large DNA fragments from Streptomyces strains. Int. J. Syst. Bacteriol., 43, 674–682.
Bryant, T.N., 2003. PIB Win-software for probabilistic identification. J. Appl. Microbiol., 97, 1326–1327.
Cook, A.E., Meyers, P.R., 2003. Rapid identification of filamentous actinomycetes to the genus level using-specific 16S rRNA gene restriction fragment patterns. Int. J. Syst. Evol. Microbiol., 53, 1907–1915.
Deng, W., Xi, D., Mao, H., Wanapat, M., 2008. The use of molecular techniques based on ribosomal RNA and DNA for rumen microbial ecosystem studies: A review. Mol. Biol. Rep., 35, 265–274.
Hankin, L., Zucker, M., Sands, D.C., 1971. Improved solid medium for the detection and enumeration of pectolytic bacteria. Appl. Microbiol., 22, 205–209.
Harvey, I., Cormier, Y., Beaulieu, C., Akimov, V.N., Meriaux, A., Duchaine, C., 2001. Random amplified ribosomal DNA restriction analysis for rapid identification of thermophilic actinomycete-like bacteria involved in hypersensitivity pneumonitis. Syst. Appl. Microbiol., 24, 277–284.
Lechevalier, M.P., Lechevalier, H.A., 1970. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Bact., 20, 435–443.
Locci, R., 1989. Streptomyces and related genera. In: Bergey’s manual of Systematic Bacteriology. Vol. IV, (Ed., Williams, ST) Baltimore: Williams and Wilkins Co., 2451–2458.
Magarvey, N.A., Keller, J.M., Bernan, V., Dworkin, M., Sherman, D.H., 2004. Isolation and characterization of Novel Marine-Derived Actinomycetes Taxa rich in bioactive metabolites. Appl. Environ. Microbial., 70, 7520–7529.
Mehling, A., Wehmeier, U.F., Piepersberg, W., 1995. Nucleotide sequences of Streptomycetes 16S ribosomal DNA: towards a specific identification system for Streptomycetes using PCR. Microbiol., 141, 2139–2147.
Nitsch, B., Kutzner, H.J., 1969. Egg-Yolk agar as diagnostic medium for Streptomycetes. Experimentia., 25, 113–116.
Schleifer, K.H., Stackebrandt, E., 1983. Molecular systematic of prokaryotes. Ann. Rev. Microbiol., 37, 143–187.
Shirling, E.B., Gottlieb, D., 1966. Methods for characterization of Streptomyces sps. Int. J. Syst. Bacteriol., 16, 313–340.
Steingrube, V.A., Brown, B.A., Gibson, J.L., Wilson, R.W., Brown, J., Blacklock, Z., Jost, K., Locke, S., Ulrich, R.F., Wallace, R.J., JR., 1995. DNA amplification and restriction endonuclease analysis for differentiation of 12 species and taxa of Nocardia, including recognition of four new taxa within the Nocardia asteroides complex. J. Clin. Microbiol., 33, 3096–3101.
Steingrube, V.A., Wilson, R.W., Brown, B.A., Jost, K.C., JR., Blacklock, Z., Gibson, J.L., Wallace, R.J., JR., 1997. Rapid identification of the clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordona, Nocardia, Rhodococcus, Streptomyces and Tsukamurella isolates by DNA amplification and restriction endonuclease analysis. J. Clin. Microbiol., 35, 817–822.
Telenti, A., Marchesi, F., Balz, M., Bally, F., Bottger, E.C., Bodmer, T., 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol., 31, 175–178.
Thakur, R., Rai, V., 2011. Isolation and biochemical identification of thermophilic Streptomyces from soil of Raipur district of Chhattisgarh, India. Res. J. Pharm. Boil. Chem. Sci., 2, 795–808.
Waksman, S.A., Henrici, A.T., 1943. The nomenclature and classification of actinomycetes. J. Bacteriol., 46, 337–341.
Williams, S.T., Shameemullah, M.J.E., Watson, T., Mayfield, C.I., 1972. Study on the ecology of actinomycetes in soil-VI. The influence of moisture tension on growth and survival. Soil. Biol. Biochem., 4, 215–225.
Wilson, R.W., Steingrube, V.A., Brown, B.A., Wallace, R.J., 1998. Clinical application of PCR-restriction enzyme pattern analysis for rapid identification of aerobic actinomycete isolates. J. Clin. Microbiol., 36, 148–152.
Zeigler, P., Kutzner, H.J., 1973. Hippurate hydrolysis as a taxonomic criterion in the genus Streptomyces (order Actinomycetales). Z. Allg. Mikrobiol., 13, 265–272.
Published
How to Cite
Issue
Section
Copyright (c) 2020 Rama Thakur, Vibhuti Rai, Varsha Shukla

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.



